linregress¶
- numeric.stats.linregress(x, y)¶
Calculate a linear least-squares regression for two sets of measurements.
- Parameters:
y (x,) – (array_like) Two sets of measurements. Both arrays should have the same length.
- Returns:
Result slope, intercept, relative coefficient, two-sided p-value for a hypothesis test whose null hypothesis is that the slope is zero, standard error of the estimated gradient, validate data number (remove NaN values).
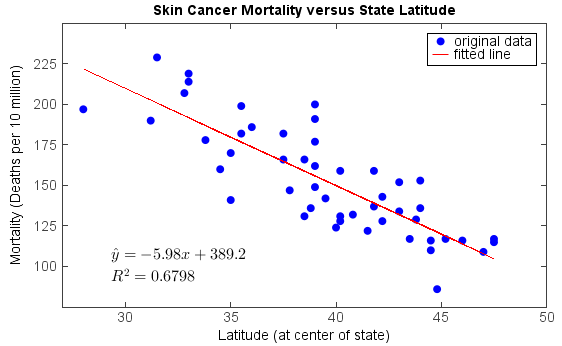
Examples:
fn = 'D:/Temp/ascii/skincancer.txt' table = readtable(fn, format='%s%f%2i%f') lat = table['Lat'] mort = table['Mort'] slope, intercept, r_value, p_value, std_err = stats.linregress(lat, mort) scatter(lat, mort, label='original data', edge=False) plot(lat, intercept + slope*lat, 'r', label='fitted line') text(29, 100, r'$\hat{y} = ' + '%.2f' % slope + 'x + ' + \ '%.1f' % intercept + '$', fontsize=16) text(29, 88, r'$R^2 = '+ '%.4f' % (r_value**2) + '$', fontsize=16) legend() xlim(27, 50) ylim(75, 250) xticks(arange(30, 51, 5)) yticks(arange(100, 226, 25)) title('Skin Cancer Mortality versus State Latitude') xlabel('Latitude (at center of state)') ylabel('Mortality (Deaths per 10 million)')